Welcome to boxtree’s documentation!¶
boxtree is a package that, given some point locations in two or three dimensions, sorts them into an adaptive quad/octree of boxes, efficiently, in parallel, using OpenCL. It also computes geometric lookup tables and generates FMM interaction lists.
Other places on the web to find boxtree stuff:
Now you obviously want to watch the library do something (at least mildly) cool? Well, sit back and watch:
import logging
import numpy as np
import pyopencl as cl
from pytools import obj_array
from boxtree import TreeBuilder
from boxtree.array_context import PyOpenCLArrayContext
from boxtree.traversal import FMMTraversalBuilder
from boxtree.visualization import TreePlotter
logging.basicConfig(level="INFO")
ctx = cl.create_some_context()
queue = cl.CommandQueue(ctx)
actx = PyOpenCLArrayContext(queue, force_device_scalars=True)
dims = 2
nparticles = 500
# -----------------------------------------------------------------------------
# generate some random particle positions
# -----------------------------------------------------------------------------
rng = np.random.default_rng(seed=15)
particles = obj_array.new_1d([
actx.from_numpy(rng.normal(size=nparticles))
for i in range(dims)])
# -----------------------------------------------------------------------------
# build tree and traversals (lists)
# -----------------------------------------------------------------------------
tb = TreeBuilder(actx)
tree, _ = tb(actx, particles, max_particles_in_box=5)
tg = FMMTraversalBuilder(actx)
trav, _ = tg(actx, tree)
This file is included in the boxtree distribution as
examples/demo.py. With some plotting code (not shown above, but
included in the demo file), you can see what’s going on:
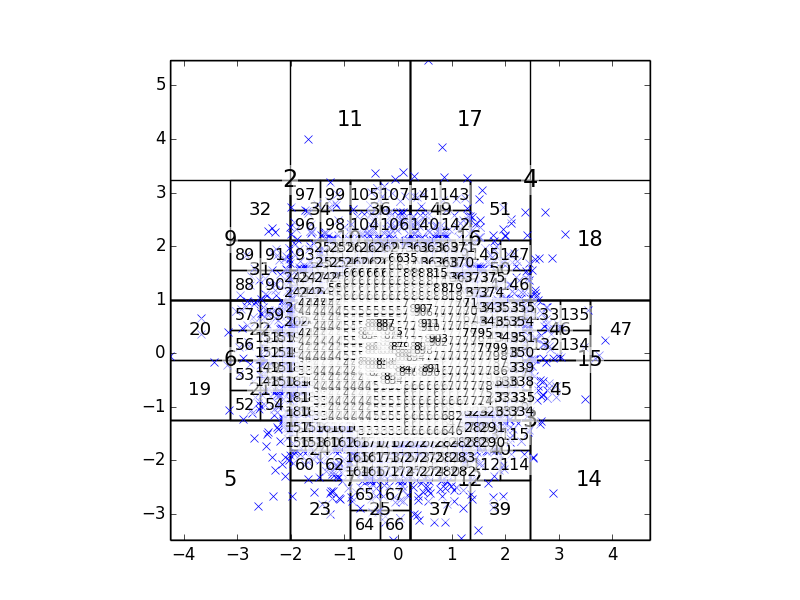
More importantly, perhaps, than being able to draw the tree, the boxtree.Tree
data structure is now accessible via the tree variable above, and the connectivity
information needed for an FMM-like traversal is available in trav as
a boxtree.traversal.FMMTraversalInfo.
Overview¶
- Introduction
- Tree Data Structures
- Building Trees
- Building interaction lists
- FMM driver
drive_fmm()TreeIndependentDataForWranglerExpansionWranglerInterfaceExpansionWranglerInterface.tree_indepExpansionWranglerInterface.traversalExpansionWranglerInterface.treeExpansionWranglerInterface.reorder_sources()ExpansionWranglerInterface.reorder_potentials()ExpansionWranglerInterface.multipole_expansions_view()ExpansionWranglerInterface.local_expansions_view()ExpansionWranglerInterface.form_multipoles()ExpansionWranglerInterface.coarsen_multipoles()ExpansionWranglerInterface.eval_direct()ExpansionWranglerInterface.multipole_to_local()ExpansionWranglerInterface.eval_multipoles()ExpansionWranglerInterface.form_locals()ExpansionWranglerInterface.refine_locals()ExpansionWranglerInterface.eval_locals()ExpansionWranglerInterface.finalize_potentials()
FMMLibTreeIndependentDataForWranglerFMMLibExpansionWrangler- Internal bits
- Tree-based geometric lookup
- FMM Cost Model
- Distributed Computation
- Utility Functionality
- Installation
- User-visible Changes
- License
- Acknowledgments
- Cross-References to Other Documentation
- 🚀 Github
- 💾 Download Releases